ASU study: Artificial cells demonstrate life's remarkable adaptability

In the study, a synthetically constructed minimal cell, derived from the bacterium M. mycoides, was reduced to a suite of essential genes. The researchers describe the functioning of a stripped-down version of this bacterial cell and its startling ability to overcome the paucity of its modified genome. Graphic by Jason Drees
A single cell is a condensed universe, containing a rich storehouse of genetic information necessary for survival and reproduction. But how essential are all those genes, and could a cell manage to flourish and evolve without so much complexity?
In new research, Michael Lynch, director of the Biodesign Center for Mechanisms of Evolution at Arizona State University, joins colleagues, including the study’s lead author Jay T. Lennon, professor of biology at Indiana University Bloomington, to explore the issue.
A synthetically constructed minimal cell, derived from the bacterium M. mycoides, was reduced to a suite of essential genes. In a surprising discovery, the researchers describe the functioning of a stripped-down version of this bacterial cell and its startling ability to overcome the paucity of its modified genome.
The team found that the streamlined cell can evolve just as fast as a normal cell — demonstrating the capacity for organisms to adapt, even with an unnatural genome that would seemingly provide little flexibility. Despite their radically simplified state, the cells displayed a robust capacity to evolve, regaining 80% of their biological fitness in around 2000 generations.
“It is not too terribly surprising that these cells were able to evolve, as evolution by natural selection is a basic property of life, but the rapidity with which improvement in cell growth rate developed in such a stream-lined genome (with fewer mutational targets than in most organisms) was certainly a surprise,” Lynch says.
The research appears in the current issue of the journal Nature.
For their study, the team used the synthetic organism Mycoplasma mycoides JCVI-syn3B — a minimized version of the bacterium M. mycoides commonly found in the guts of goats and similar animals. Over millennia, the parasitic bacterium has naturally lost many of its genes as it has evolved to depend on its host for nutrition.
In previous research, scientists at the J. Craig Venter Institute in California took this one step further. In 2016, they eliminated 45% of the 901 genes from the natural M. mycoides genome, reducing it to the smallest set of genes required for autonomous cellular life. At 493 genes, the minimal genome of M. mycoides JCVI-syn3B is the smallest of any known free-living organism. In comparison, many animal and plant genomes contain more than 20,000 genes.
In principle, the simplest organism would have no functional redundancies and possess only the minimum number of genes essential for life. Any mutation in such an organism could lethally disrupt one or more cellular functions, placing constraints on evolution. Organisms with streamlined genomes have fewer targets upon which positive selection can act, thus limiting opportunities for adaptation.
Michael Lynch
Although M. mycoides JCVI-syn3B could grow and divide in laboratory conditions, the researchers wanted to know how a minimal cell would respond to the forces of evolution over time, particularly given the limited raw materials upon which natural selection could operate as well as the uncharacterized input of new mutations.
In the M. mycoides JCVI-syn3B minimalist cells, every single gene in its genome is essential. This might, in principle, leave little room for exploratory adaptive evolution or beneficial mutations, possibly constraining the potential to evolve.
Instead, the researchers established that M. mycoides JCVI-syn3B has an exceptionally high mutation rate. They then grew it in the lab where it was allowed to evolve freely for 300 days, equivalent to 2,000 bacterial generations or about 40,000 years of human evolution.
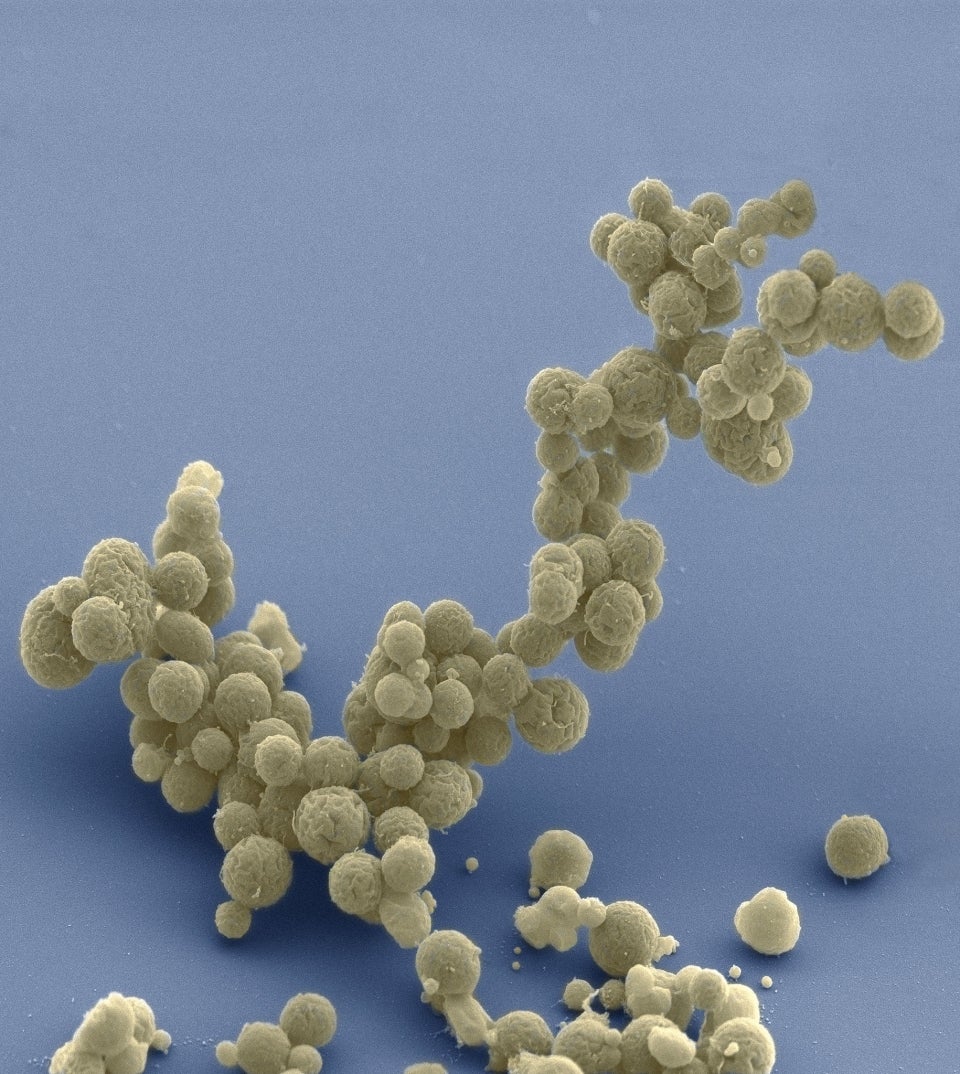
Electron micrograph of a cluster of minimal cells magnified 15,000 times. The synthetically streamlined bacterium Mycoplasma mycoides contains less than 500 genes. Image courtesy Tom Deerinck and Mark Ellisman of the National Center for Imaging and Microscopy Research at the University of California at San Diego
The next step was to set up experiments to determine how the minimal cells that had evolved for 300 days performed in comparison with the original, nonminimal M. mycoides, as well as with a strain of minimal cells that hadn't evolved for 300 days. In the comparison tests, the researchers put equal amounts of the strains being assessed together in a test tube. The strain better suited to its environment became the more common strain.
They found that the nonminimal version of the bacterium easily outcompeted the unevolved minimal version. The minimal bacterium that had evolved for 300 days, however, did much better, effectively recovering all of the fitness that it had lost due to genome streamlining. The researchers identified the genes that changed the most during evolution. Some of these genes were involved in constructing the surface of the cell, while the functions of several others remain unknown.
Understanding how organisms with simplified genomes overcome evolutionary challenges has important implications for long-standing problems in biology — including the treatment of clinical pathogens, the persistence of host-associated endosymbionts, the refinement of engineered microorganisms, and the origin of life itself.
The research carried out by Lynch and his team demonstrate the power of natural selection to rapidly optimize fitness in the simplest autonomous organism, with implications for the evolution of cellular complexity.
More Science and technology

ASU water polo player defends the goal — and our data
Marie Rudasics is the last line of defense.Six players advance across the pool with a single objective in mind: making sure that…

Diagnosing data corruption
You are in your doctor’s office for your annual physical and you notice the change. This year, your doctor no longer has your…
Large-scale study reveals true impact of ASU VR lab on science education
Students at Arizona State University love the Dreamscape Learn virtual reality biology experiences, and the intense engagement it…