Comprehensive statewide study tracks rapid spread of SARS-CoV-2 variants

Efrem Lim is a researcher in the Biodesign Center for Fundamental and Applied Microbiomics and an associate professor with the ASU School of Life Sciences.
Having carved a path of destruction around the globe, the novel coronavirus SARS-CoV-2 continues to recur in ever-changing disguises. Understanding the dynamics of viral transmission is crucial for ongoing and future public health preparedness.
In a first-of-its-kind, comprehensive statewide survey, Efrem Lim and his colleagues at Arizona State University’s Biodesign Institute teamed up with three regional hospitals to study the spread of SARS-CoV-2 variants.
The study will help researchers, clinicians and public health officials better understand how quickly variants of the virus rise in prevalence throughout Arizona. The lightning-fast spread of the omicron variant is explored in detail through surveillance of community testing, hospital data and sequencing of viral variants found in community wastewater.
The findings highlight the fact that SARS-CoV-2 underwent a substantial alteration of its genomic profile in its transformation to omicron, a phenomenon known as antigenic shift. The emergence rate of omicron BA.1 (17.02 days) was over 2.3 times faster than the previous delta variant and the subsequent omicron lineages BA.2 and BA.5.
Lim is a researcher in the Biodesign Center for Fundamental and Applied Microbiomics and an associate professor with the ASU School of Life Sciences. The research incorporates community sequencing information from public clinics as well as from three hospitals located in the Phoenix metropolitan area: Valleywise Health, Phoenix Children’s and Dignity Health.
The study follows the omicron variant, observing its impact on hospital admissions and comparing this data with viral strains extracted from wastewater. Having analyzed the rapid ascent of omicron BA.1, “we now have a benchmark for all these other variants that are coming up,” Lim says. “Are they rising as fast as the superstar, the omicron BA.1? If they are, then that's a red flag that we need to start preparing for a very large surge, like the omicron wave.”
The research appears in the current issue of the American Society for Microbiology journal mBIO.
Shift in the viral strains
An antigenic shift is a sudden change in the genetic makeup of a virus or other pathogen, which leads to the creation of a new strain that is significantly different from previous strains. This can occur through mutation or through the reassortment of genetic material from different strains of the virus.
Such a shift in viral character can have consequential implications for public health, as the new strain may be able to evade the immunity developed by individuals who have been previously infected or vaccinated against the original strain of the virus, leading to more widespread outbreaks.
“There’s a difference between what we call antigenic shift versus antigenic drift,” Lim says.
A good example is the influenza virus, which typically undergoes small, incremental changes from year to year, requiring the production of new annual vaccines.
“But occasionally, the virus undergoes very large changes such as recombination with other influenza viruses like avian influenza or swine influenza. This is what we call an antigenic shift, and whenever a virus undergoes antigenic shift, it always leads to a large pandemic,” Lim says.
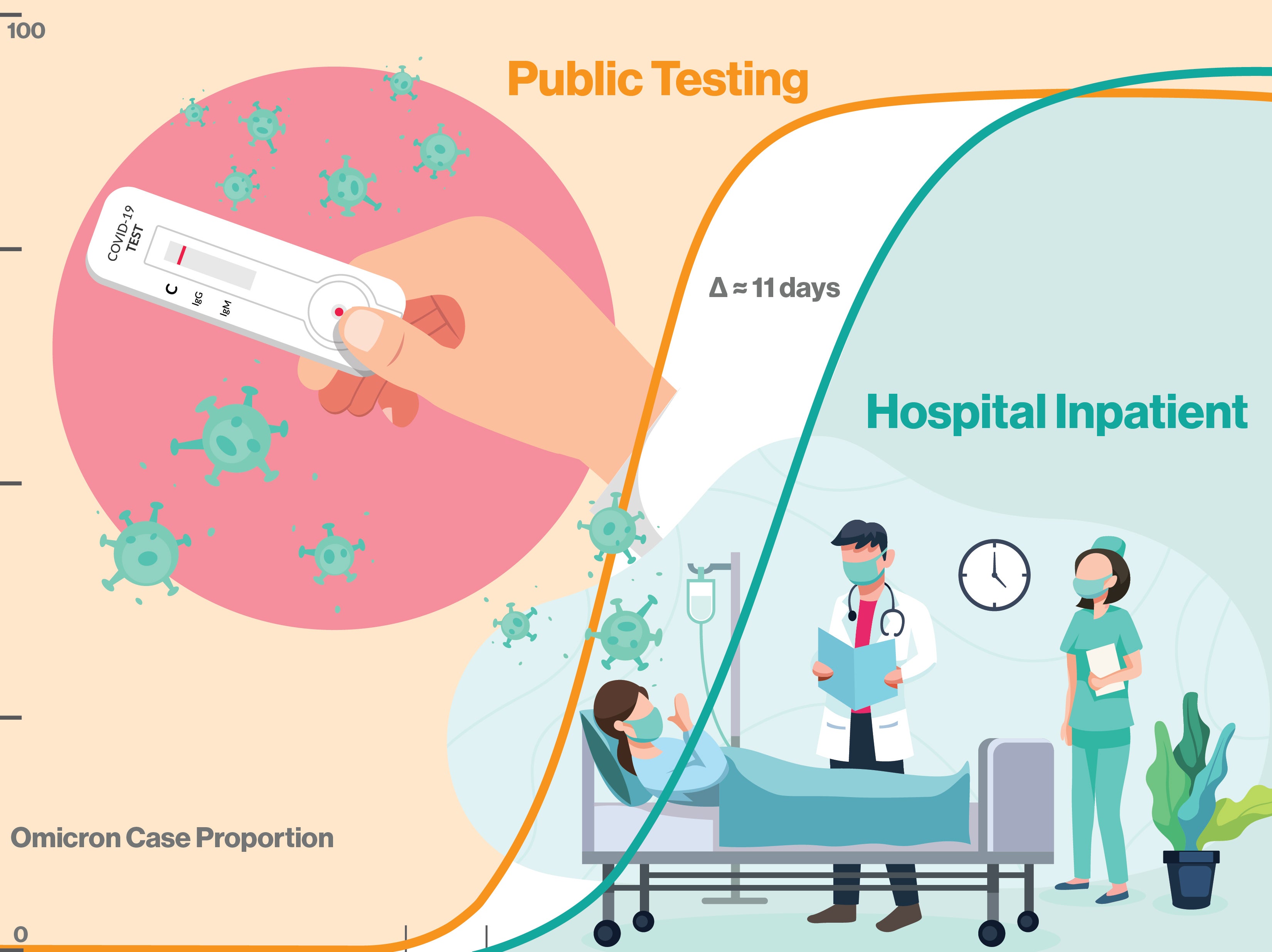
The emergence rate of of SARS-CoV-2 variant omicron BA.1 was over 2.3 times faster than the previous delta variant and the subsequent omicron lineages BA.2 and BA.5. Fewer than 11 days elapsed from the BA.1 variant's first community appearance to subsequent hospitalizations. The BA.1 variant can now act as a benchmark for tracking the appearance and spread of new variants, giving public health officials and hospitals advanced warning of emergent variants posing the highest risk to the community. Graphic by Shireen Dooling/ASU
Influenza pandemics like the 2009 H1N1 outbreak were the result of antigenic shifts. The same rules apply for the SARS-CoV-2 virus.
The study explored the genomic epidemiology of SARS-CoV-2 at the state level by conducting baseline genomic sequencing surveillance of more than 27,000 samples from public testing and 1,125 samples from hospitalized patients, which were diagnosed between Nov. 1, 2021, and Jan. 31, 2022, in Arizona. Results indicate that omicron cases were identified in hospitals just 10.51 days after the variant’s first emergence in the community.
After omicron emergence and a brief lag of around 10 days, it caused a surge in hospital admissions, as the study verified. Researchers also identified mutations in three omicron genes that were significantly more common in cases of COVID-19 requiring hospitalization, compared with positive COVID-19 cases from community testing.
First-time sequencing of SARS-CoV-2 variants from wastewater
Wastewater-based epidemiology is a new and effective public health approach to monitoring communitywide infectious diseases occurrence. The advantages of wastewater monitoring during the COVID-19 pandemic include estimating disease burden despite decreased testing rates in the community over time. Presymptomatic and asymptomatic infections that may not normally be tested can also be evaluated through wastewater sequencing, identifying the viral lineages circulating within a community.
The study marks the first-time sequence data pertaining to variants of SARS-CoV-2 have been extracted from wastewater, significancy extending the power of the technique for public health surveillance. Wastewater monitoring and clinical surveillance are complementary approaches to public health efforts. In recent months, community and wastewater data on viral presence have diverged, suggesting that while cases of the virus are still rising, reduced testing habits in the community may be failing to register many of these.
One important lesson from studying the rise of omicron BA.1 is the tiny available window between the emergence of new variants and the first hospital cases. As omicron BA.1 demonstrates, this can occur in as little as 10 days, with the possibility of overwhelming underprepared hospitals. While most viral alterations will be minor and incremental, the chance of antigenic shift and sudden, heightened disease incidence remain a serious challenge for public health. Integrative, cooperative efforts, leveraging the resources of hospitals, community data and new sequencing methods will be essential as humanity continues to grapple with SARS-CoV-2 and other infectious threats.
More Science and technology

ASU to host 2 new 51 Pegasi b Fellows, cementing leadership in exoplanet research
Arizona State University continues its rapid rise in planetary astronomy, welcoming two new 51 Pegasi b Fellows to its exoplanet…

ASU students win big at homeland security design challenge
By Cynthia GerberArizona State University students took home five prizes — including two first-place victories — from this year’s…

Swarm science: Oral bacteria move in waves to spread and survive
Swarming behaviors appear everywhere in nature — from schools of fish darting in synchrony to locusts sweeping across landscapes…