ASU researchers unraveling protein structure to understand and fight disease

Abhishek Singharoy is a researcher with ASU's Biodesign Center for Applied Structural Discovery and an assistant professor with the School of Molecular Sciences.
Proteins are one of the major building blocks of life, and they carry out many chemical reactions related to life. This includes how the body interacts with disease-causing agents. Understanding and treating diseases requires scientists to determine the structures and shapes of proteins involved in these interactions.
Three-dimensional computer modeling and electron microscopy are two of the tools scientists are currently using to help learn more about these proteins. To spur development of more effective tools, EMDataResource, a global organization for the storage and retrieval of three-dimensional electron microscopy data, sponsored the 2019 Cry0-EM Map-based Model Metrics Challenge. The goals of this challenge were to assess the quality of models that can be produced using current modeling software; check the reproducibility of modeling results from different software developers and users; and compare the performance of current metrics used for evaluation of models.
Abhishek Singharoy, assistant professor with Arizona State University's School of Molecular Sciences and researcher with ASU's Biodesign Center for Applied Structural Discovery, led the CryoFold team. Singharoy’s group was recognized in the challenge for doing very well in both lower- and higher-resolution targets. CryoFold is an Ab-initio modeling platform interfacing between three popular software tools such as MAINMAST, developed by Genki Terashi and Professor Daisuke Kihara at Purdue University; Modeling Employing Limited Data (MELD), developed by Alberto Perez, now at University of Florida, and Professor Ken Dill, Stony Brook University; and Resolution Exchange Molecular Dynamics Flexible Fitting (ReMDFF) developed by Singharoy and Professor Klaus Schulten, University of Illinois at Urbana-Champaign.
Singharoy noted: “The work assessed by members of Protein Data Bank (PDB) and Electron-Microscopy Data Bank (EMDB) highlights the importance of integrative modeling in the fields of structural biology, biophysics and biochemistry. It is very encouraging for our team members to see the excellent performance of molecular dynamics flexible fitting (MDFF) methods in this competition.”
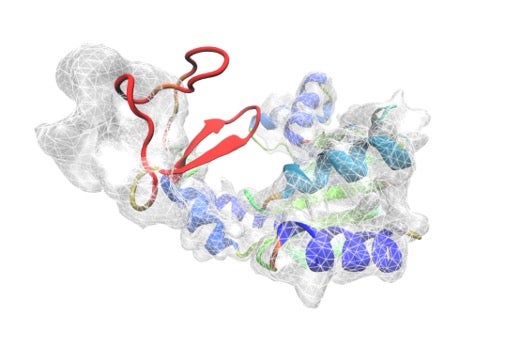
Electron density map revealed by cryo-EM techniques, showing peptide geometry of a protein.
“It is exciting to see the advances and successes coming from combining cryoEM maps and computational tools to provide atomistic detail of proteins. Solving the protein structure is crucial to understanding how they work and to design new drugs that target them,” said Perez.
A paper on the results of the competition is published in the journal Nature Methods.
Singharoy offered, “Understanding three-dimensional protein structures will help us tackle many chronic and fatal diseases associated with proteins.”
More Science and technology

ASU at the heart of the state's revitalized microelectronics industry
A stronger local economy, more reliable technology, and a future where our computers and devices do the impossible: that’s the…

Breakthrough copper alloy achieves unprecedented high-temperature performance
A team of researchers from Arizona State University, the U.S. Army Research Laboratory, Lehigh University and Louisiana State…

4 ASU researchers named senior members of the National Academy of Inventors
The National Academy of Inventors recently named four Arizona State University researchers as senior members to the prestigious…

