ASU professor tackles life at the atomic level with NSF CAREER award

Assistant Professor Abhishek Singharoy has just been awarded a NSF CAREER award. Photo by Mary Zhu
Assistant Professor Abhishek Singharoy from Arizona State University’s School of Molecular Sciences and the Biodesign Institute’s Center for Applied Structural Discovery has recently earned a Faculty Early Career Development (CAREER) award from the National Science Foundation (NSF).
The prestigious CAREER program supports the early career development activities of teacher-scholars who most effectively integrate research and education within the mission of their organization. It provides five-year research grants to each recipient.
“In our Systems Structural Biology Group we make a computational microscope, which allows us to collect experimental data and combine it with huge computer simulations to show us life in its greatest possible detail,” Singharoy explained. “With this computational microscope, one can start learning about energy and how life uses energy and processes energy to convert it from one form to another.”
“Abhishek Singharoy and his group are doing extremely innovative molecular science, developing new approaches to achieve detailed molecular understanding of more complex chemical and biochemical systems than have ever been modeled previously,” said Neal Woodbury, director of the School of Molecular Sciences. “Our younger faculty members in the School of Molecular Sciences have an extraordinary record of achievement, and professor Singharoy is an exemplar in this regard.”
High school biology textbooks tell us about mitochondria being a part of all eukaryotic cells, and that the mitochondrial membrane is the powerhouse of the cell. It converts energy from the food we eat into energy that runs a large range of biological processes.
However very little is currently known about how this energy conversion occurs at the atomic level; this is what is being probed by Singharoy and his group. He seeks to understand the chemistry of the electron and how it translates into cell function.
In order to study what is happening at the nanoscale, Singharoy employs both experimental studies (e.g. cryogenic electron microscopy) and computer simulations on a massive scale (100 million atoms). The simulations take advantage of nanoscale molecular dynamics and visual molecular dynamics at the Summit supercomputing facilities at Oak Ridge National Lab in Tennessee. The computational work also involves a collaboration with the University of Illinois Urbana Champagne, Purdue University and the University of Pittsburgh.

Structural Systems Biology Group (from left to right): Jose Garcia (Fromme group), Abhishek Singharoy, Michael Moran (Fromme group), Eric Wilson, Jon Nguyen, John Vant, Chitrak Gupta, Steven Razo and Jacob Layton.
Single-particle cryogenic electron microscopy allows researchers to take a look at biological structures that were simply not accessible just a few years ago, and it is now exposing structures of unprecedented complexity in great detail. ASU is the perfect place for Singharoy to conduct these studies: The Titan electron microscope at ASU is capable of exceptional atomic-resolution imaging of materials in high-vacuum. The other two essential components at ASU are the Agave computer cluster and a collaboration with Professor Wayne Frasch in the School of Life Sciences. Frasch and doctoral student Seiga Yanagisawa have developed new assays to examine the rotation of single molecules of molecular motor proteins under a microscope.
John Vant and Jon Nguyen, Singharoy’s doctoral students, are studying the charge transport chain in mitochondria. Cytochrome c is a small hemeprotein associated with mitochondria, and electrons are efficiently shuttled between complex III and complex IV — both cytochrome c enzymes. The study focuses on dissecting the evolutionary design principles of mitochondrial respiration, in particular, through investigation of ATPases and the outer membrane-embedded supercomplexes.
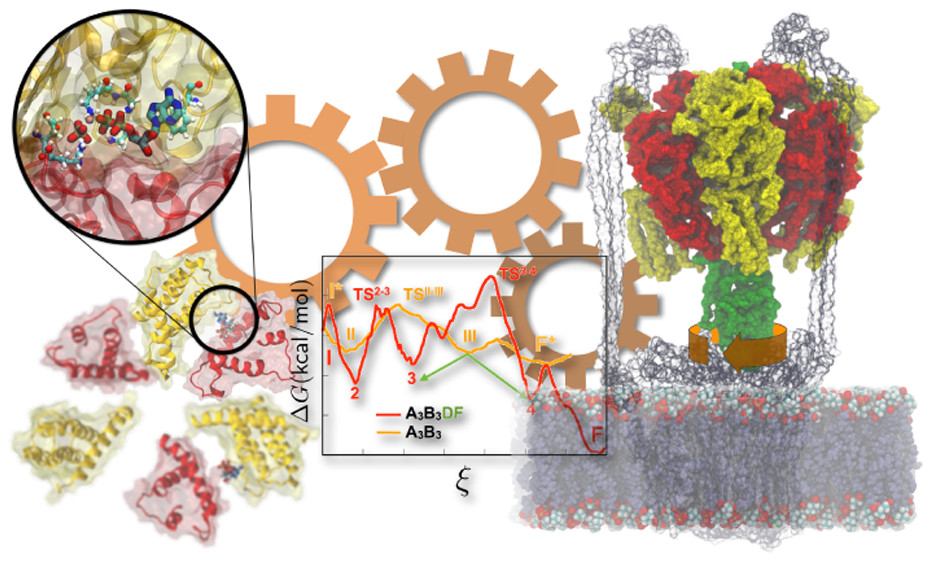
Supercomplex proteins embedded in the outermitochondrial membrane.
In the course of the massive computer studies, Singharoy’s group ended up developing some software using a molecular dynamics simulation tool that they call NAMD. Some of this software is now freely available online on cloud resources, connecting high school libraries and bringing free biophysics education to high schools in Arizona and beyond.
“Students can start interacting with our computers from their libraries and learn more about proteins and waters and membranes, while they’re studying their textbooks to give a complete picture of how science looks, both inside and out,” Singharoy said.
The project brings to light a couple of cutting-edge biomedical applications, namely, determination of the molecular origins of cellular aging and programmed cell death, and creation of a novel computer-aided pipeline pertaining to intricate pathology of the respiratory network. To put together large-scale membrane systems in atomic detail requires theoretical advances in terms of fitting/refining structural data from experiments. To address this need, group members have been developing and applying an array of flexible-fitting tools that derive high-resolution molecular models from low-resolution experimental data, such as from X-ray crystallography, electron microscopy and quantitative mass-spectrometry.
More Science and technology

4 ASU researchers named senior members of the National Academy of Inventors
The National Academy of Inventors recently named four Arizona State University researchers as senior members to the prestigious…

Transforming Arizona’s highways for a smoother drive
Imagine you’re driving down a smooth stretch of road. Your tires have firm traction. There are no potholes you need to swerve to…

The Sun Devil who revolutionized kitty litter
If you have a cat, there’s a good chance you’re benefiting from the work of an Arizona State University alumna. In honor of…

