ASU research collaboration is expanding design strategies in structural DNA nanotechnology

Assistant Professor Petr Sulc (left) and grad research assistant Erik Poppleton.
Arizona State University’s Assistant Professor Petr Sulc of the School of Molecular Sciences and the Biodesign Institute recently collaborated with Assistant Professor Thorsten L. Schmidt of Kent State, formerly of Technische Universität Dresden, Germany, to study structural DNA nanotechnology.
The field of DNA nanotechnology has been one of the most rapidly growing areas of study. Researchers are able to construct intricate DNA sequences, while having precise control over the assembled features. The uses for these applications could yield cost-effective solutions in nanomedicine, and DNA nanotechnology could change the way diseases like cancer or cardiovascular disease are diagnosed or treated in the future.
In a recent publication "Triangulated Wireframe Structures Assembled Using Single-Stranded DNA Tiles" in ACS Nano, Sulc's group developed new high-performance simulations to study DNA nanomaterials developed in Schmidt's group in TU Dresden. These novel "DNA triangulated wireframe tiles" allow for assembly of 2D and 3D nanoscale objects out of single-stranded DNAs. These objects assemble out of DNA strands which form a triangular mesh, designed to assemble into a desired shape. However, it turned out that certain structures assembled with no or very low yield. Coarse-grained model and tools for high-performance computation on GPUs, developed in the Sulc lab, were able to simulate the nanostructures consisting of several thousands of nucleotides, and explain at the molecular interaction level the reason for successful or unsuccessful assembly of the designed nanoarrays.
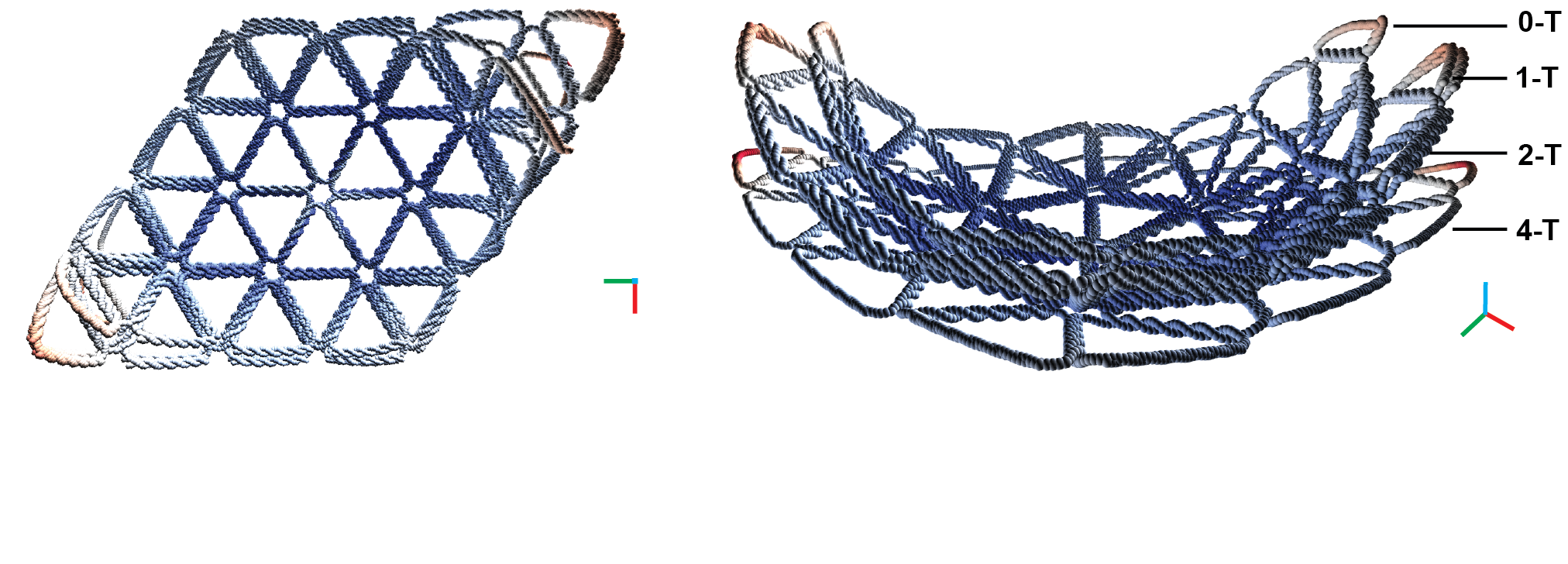
Coarse-grained molecular dynamics simulations using the oxDNA model of the rhombus structures (left) and a side view showing combined modeled structures showing distinct curvature (right).
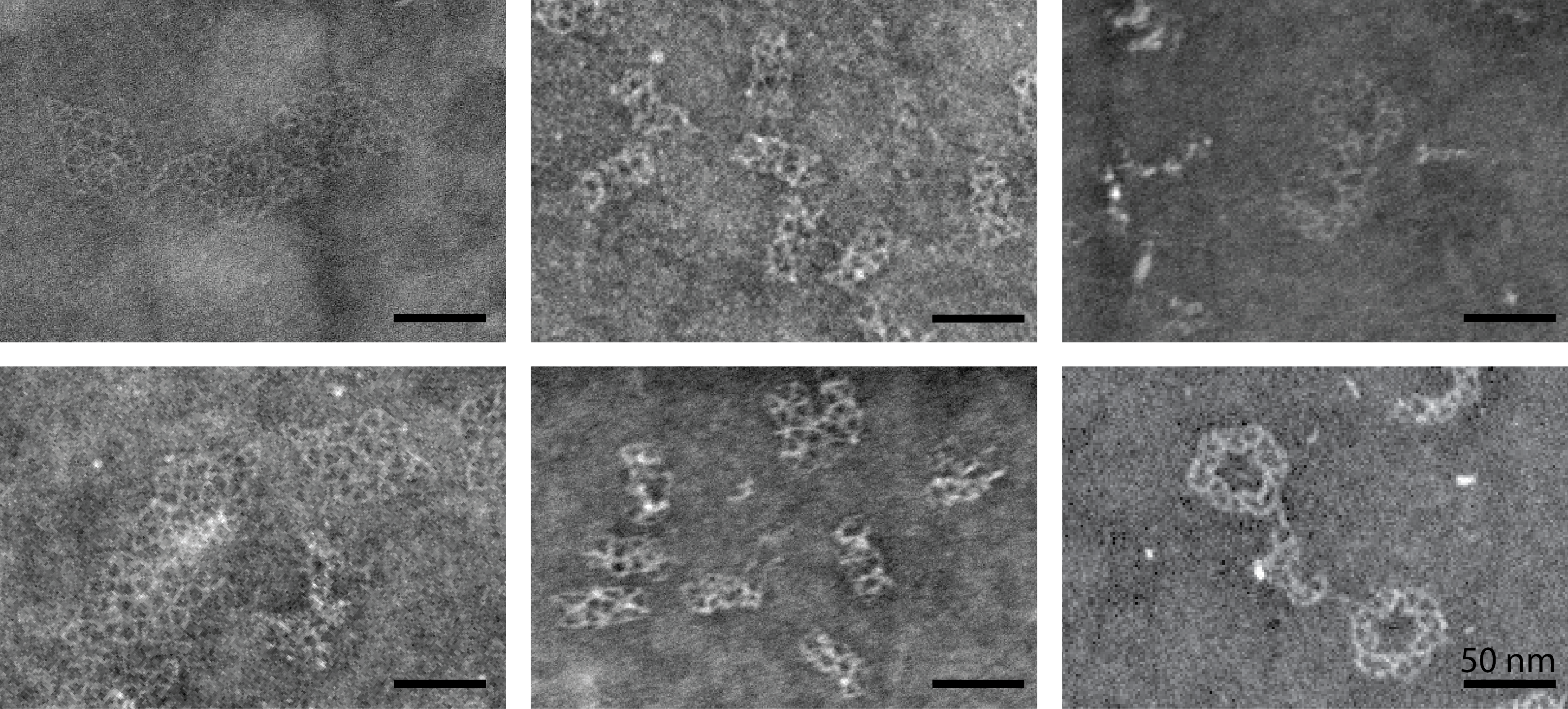
Structural analysis of the rhombus, tube and ring structures.
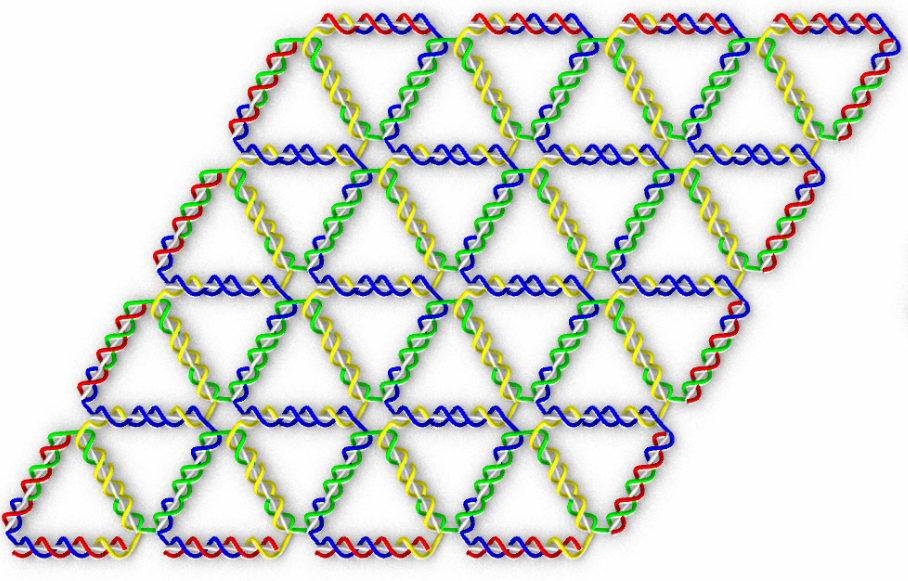
Schematics of the simulation and experiment from Sulc's work.
Sulc's lab applies broadly the methods of statistical physics and computational modeling to problems in biology, chemistry and nanotechnology. He develops new multiscale models to study interactions between biomolecules. Most of his current efforts focus on modeling nucleic acid interactions in DNA/RNA nanotechnology.
Sulc joined the School of Molecular Sciences and the Center for Molecular Design and Biomimetics at the Biodesign Institute in January 2018. The highly collaborative and interdisciplinary environment at ASU allows him to directly work side-by-side with experimental groups and develop new computer tools and algorithms that can address challenging problems that arise from nanotechnology, as well as use computer modeling to design novel nanostructures that guide experimental design.
Additional co-authors of this paper include Erik Poppleton from the Biodesign Center for Molecular Design & Biomimetics; Nayan P. Agarwal, Foram M. Joshi and Michael Matthies from the Center for Advancing Electronics Dresden, Faculty of Chemistry and Food Chemistry.
More Science and technology

Will this antibiotic work? ASU scientists develop rapid bacterial tests
Bacteria multiply at an astonishing rate, sometimes doubling in number in under four minutes. Imagine a doctor faced with a…

ASU researcher part of team discovering ways to fight drug-resistant bacteria
A new study published in the Science Advances journal featuring Arizona State University researchers has found…

ASU student researchers get early, hands-on experience in engineering research
Using computer science to aid endangered species reintroduction, enhance software engineering education and improve semiconductor…