X-ray pulses reveal structure of viral cocoon
Members of the Biodesign Center for Applied Structural Discovery.
Arizona State University’s Biodesign Center for Applied Structural Discovery (BCASD) and an international team of scientists have used high-intensity X-ray pulses to determine the structure of the crystalline protein envelope of an insect virus. Their analysis reveals the fine details of the building blocks that make up the viral cocoon down to a scale of 0.2 nanometers (millionths of a millimeter) — approaching atom-scale resolution.
The tiny viruses with their crystal casing are by far the smallest protein crystals ever analyzed using X-ray crystallography. This opens up new opportunities in the study of protein structures, as the team headed by DESY’s Leading Scientist Henry Chapman from the Center for Free-Electron Laser Science reports in the Proceedings of the National Academy of Sciences (PNAS).
“The granulovirus attacks certain insects and kills them. This initially leaves it stranded inside the decaying host, so it has to protect itself, perhaps for years, against adverse environmental conditions such as heat, ultraviolet radiation and drought, until it is once again ingested by an insect. To achieve this, the virus wraps itself in a cocoon made of protein crystals, which only dissolve again once it reaches an insect’s gut,” explained Cornelius Gati from DESY, the lead author of the paper.
The researchers examined the cocoon of the Cydia pomonella granulovirus (CpGV), which infects the caterpillars of the codling moth (Cydia pomonella) and is used in agriculture as a biological pesticide. The virus is harmless to humans.
Petra Fromme, study co-author and director of BCASD, notes the rapid pace of advance in structural discovery, owing to the improved ability of researchers to produce high-quality nanocrystals and more precisely analyze the resulting diffraction patterns produced when samples are subjected to the brilliant X-ray bursts delivered by X-ray Free Electron Laser (XFEL) instruments.
“So many important biological processes, like how we get sick or how plants capture sunlight, have been very difficult to study at the molecular level,” said Fromme. “This is a beautiful example of nature providing a clue to help scientist build better and smaller nanocrystals that will ultimately help us solve relevant and important problems in our world.”
In 2011, Fromme, along with ASU professor of physics John Spence and their collaborators, pioneered a method known as serial femtosecond crystallography. The technique is used to examine thousands of nanocrystals, using X-ray pulses at the femtosecond scale, (a millionth of a billionth of a second). Although the nanocrystals are vaporized by the X-ray pulse, the time frame is so short that a diffraction image is obtained before sample destruction occurs.
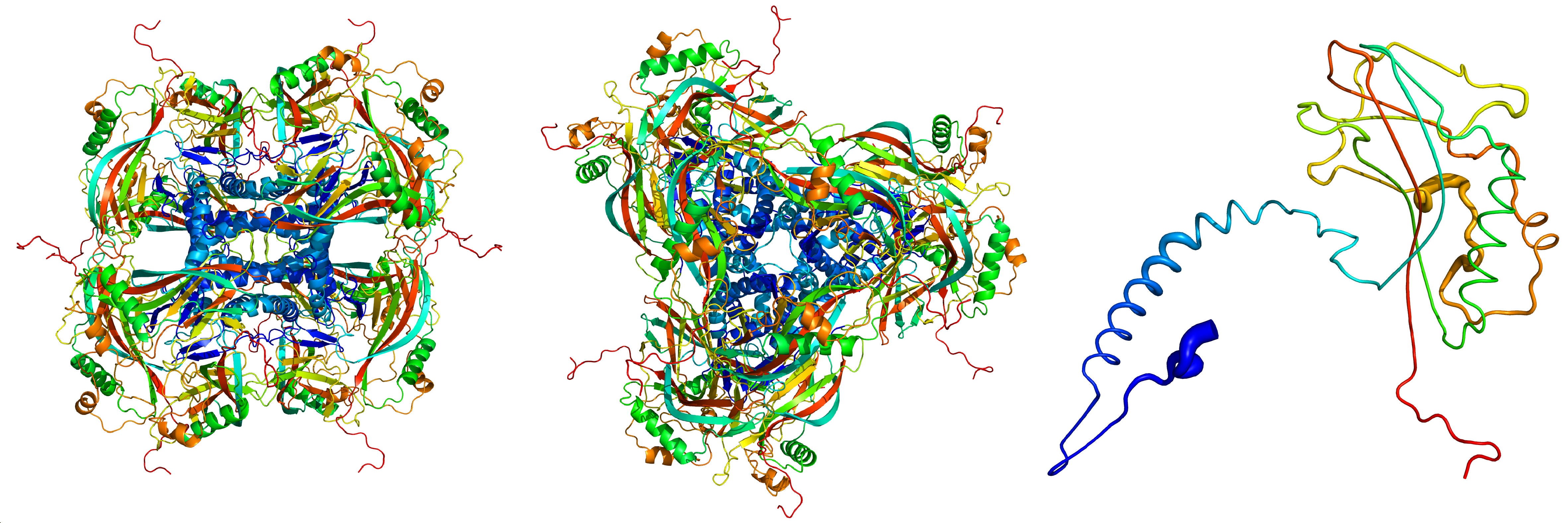
Atomic model of the crystalline occlusion bodies, derived from the X-ray diffraction images recorded at the LCLS. The individual proteins (right) stick together to form the building blocks (left, seen from the side; center, seen from above) of the crystalline occlusion bodies. Credit: Dominik Oberthür/CFEL/DESY
“With the success of this new study, we have made a major stride toward the goal of analyzing individual molecules,” said Spence, who also participated in the new work, along with fellow ASU researchers Nadia A. Zatsepin, Uwe Weierstall, R. Bruce Doak, Raimund Fromme, Ingo Grotjohann, Shibom Basu, Daniel James, Christopher Kupitz, Kimberly Rendek and Dingjie Wang.
Scientists are interested in the spatial structure of proteins and other biomolecules because this sheds light on the precise way in which they work. This has led to a specialized science known as structural biology.
“Over the past 50 years, scientists have determined the structures of more than 100,000 proteins,” said Chapman, who is also a professor of physics at the University of Hamburg. “By far the most important tool for this is X-ray crystallography.”
One challenge, however, has been producing nanocrystals appropriate for this research. Many proteins do not readily align to formcrystals, because that is not their natural state. The smaller the crystals that can be used for the analysis, the easier it is to grow them, but the harder it is to measure them.
“We are hoping that in the future we will be able to dispense altogether with growing crystals and study individual molecules directly using X-rays,” said Chapman, “so we would like to understand the limits.”
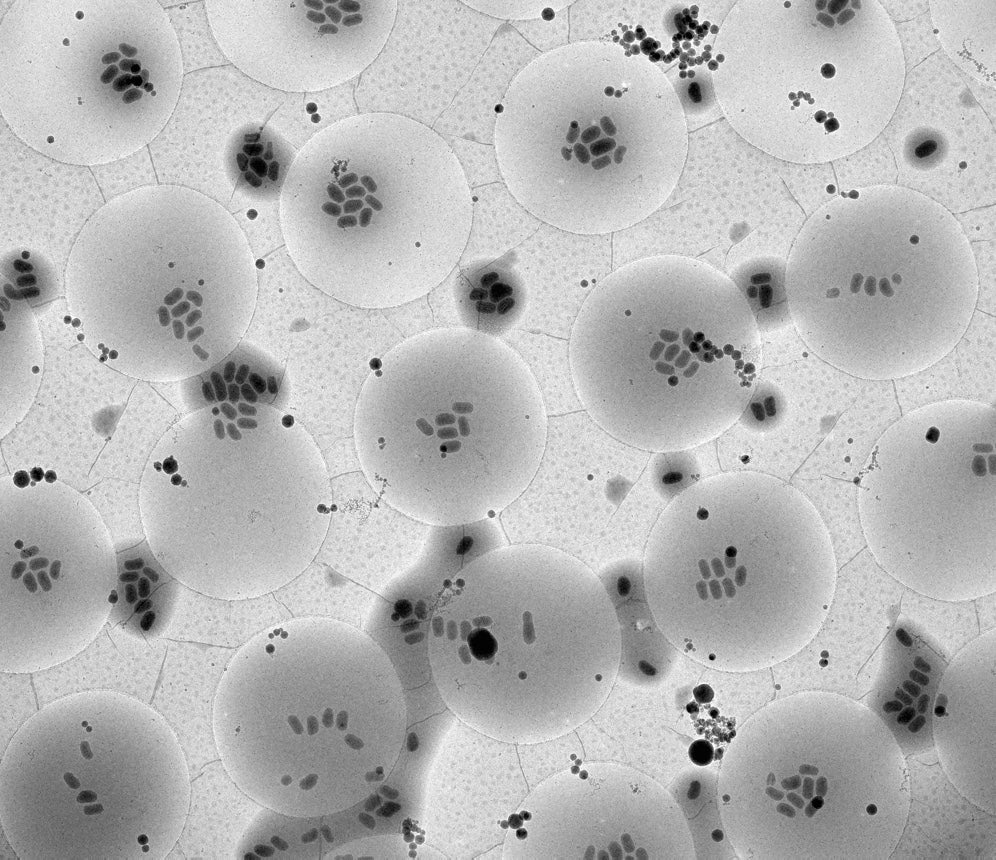
Transmission electron microscope image of granulovirus particles on an object grid. The field of view measures approximately 50 microns (0,05 millimeters). Credit: Ken Goldie/C-CINA Basel
The virus particles used in the new study provided the smallest protein crystals ever used for X-ray structure analysis. The occlusion body (the virus “cocoon”) has a volume of around 0.01 cubic micrometers, about 100 times smaller than the smallest artificially grown protein crystals that have until now been analyzed using crystallographic techniques.
To break this limit in crystal size, an extremely bright X-ray beam was needed, which was obtained using an XFEL, in which a beam of high-speed electrons is guided through a magnetic undulator causing them to emit laser-like X-ray pulses.
The scientists used the free-electron laser Linac Coherent Light Source (LCLS) at the SLAC National Accelerator Laboratory in the U.S., and employed optics to focus each X-ray pulse to a similar size as one of the virus particles. Directing the entire power of the FEL onto one tiny virus exposed it to the tremendous radiation levels, equivalent to 1.3 billion Grays. (For comparison: The lethal dose for humans is around 50 Grays.)
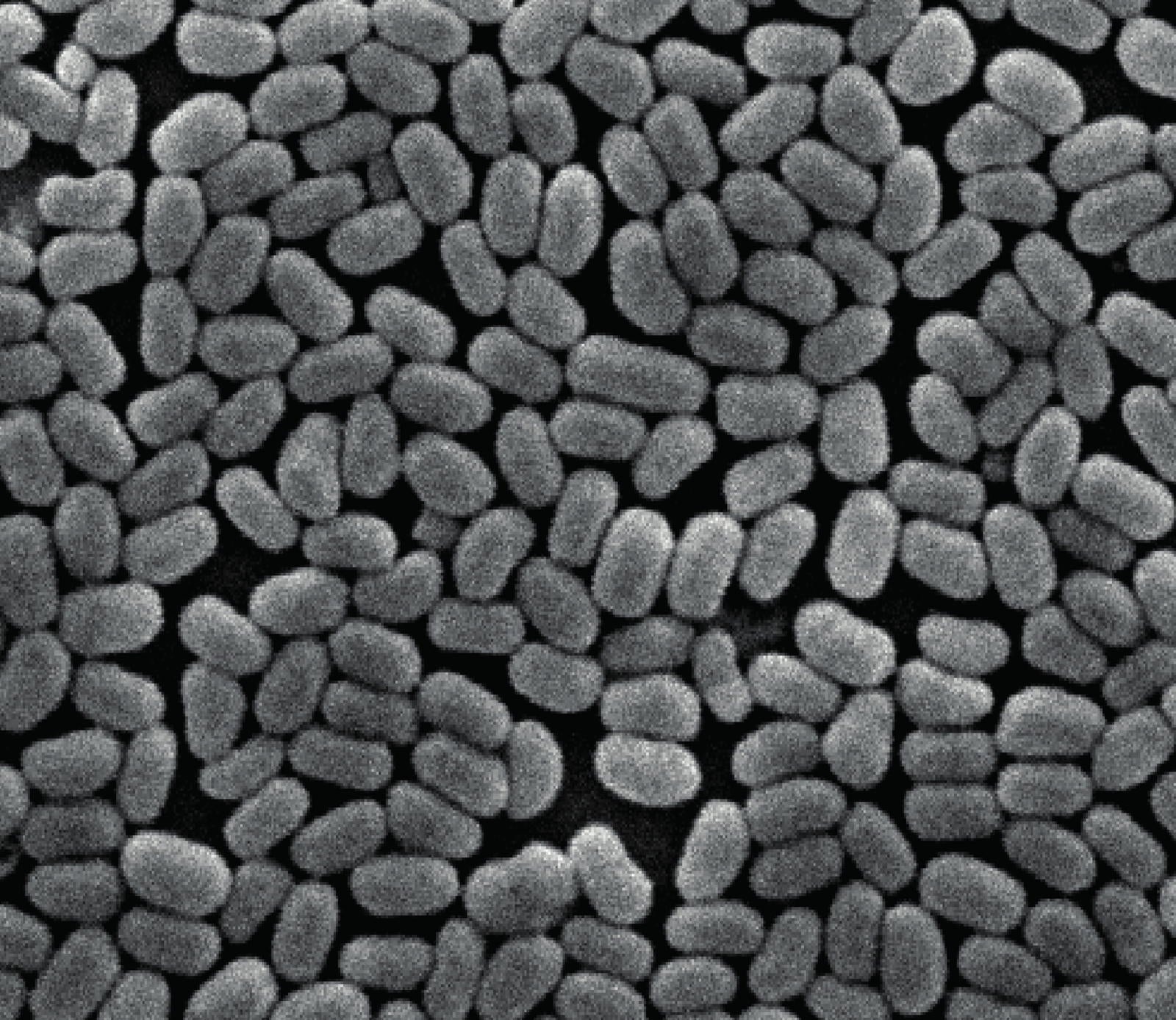
Scanning electron microscope image showing the homogeneous size of the virus particles. Field of View is about 5 microns. Credit: Peter Metcalf/University of Auckland
The FEL dose was certainly lethal for the viruses too — each was completely vaporized by a single X-ray pulse. But the femtosecond-duration pulse carries the information of the pristine structure to the detector and the destruction of the virus occurs only after the passage of the pulse. The analysis of the recorded diffraction showed that even tiny protein crystals that are bombarded with extremely high radiation doses can still reveal their structure on an atomic scale.
“Simulations based on our measurements suggest that our method can probably be used to determine the structure of even smaller crystals consisting of only hundreds or thousands of molecules,” reported Chapman, who is also a member of the Hamburg Center for Ultrafast Imaging (CUI).
Reference:
Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser; Cornelius Gati et al.; PNAS, 2017; DOI: 10.1073/pnas.1609243114
Contact info:
Petra Fromme
Center for Applied Structural Discovery, The Biodesign Institute, School of Molecular Sciences
Phone: 1 480 965 9028
E-mail: pfromme@asu.edu
Joseph Caspermeyer
Managing Editor: Biodesign Institute
p: (480) 727-0369
Joseph.Caspermeyer@asu.edu
More Science and technology

ASU researcher part of team discovering ways to fight drug-resistant bacteria
A new study published in the Science Advances journal featuring Arizona State University researchers has found…

ASU student researchers get early, hands-on experience in engineering research
Using computer science to aid endangered species reintroduction, enhance software engineering education and improve semiconductor…

ASU professor honored with prestigious award for being a cybersecurity trailblazer
At first, he thought it was a drill.On Sept. 11, 2001, Gail-Joon Ahn sat in a conference room in Fort Meade, Maryland.…