Fast, low-cost DNA sequencing technology one step closer to reality
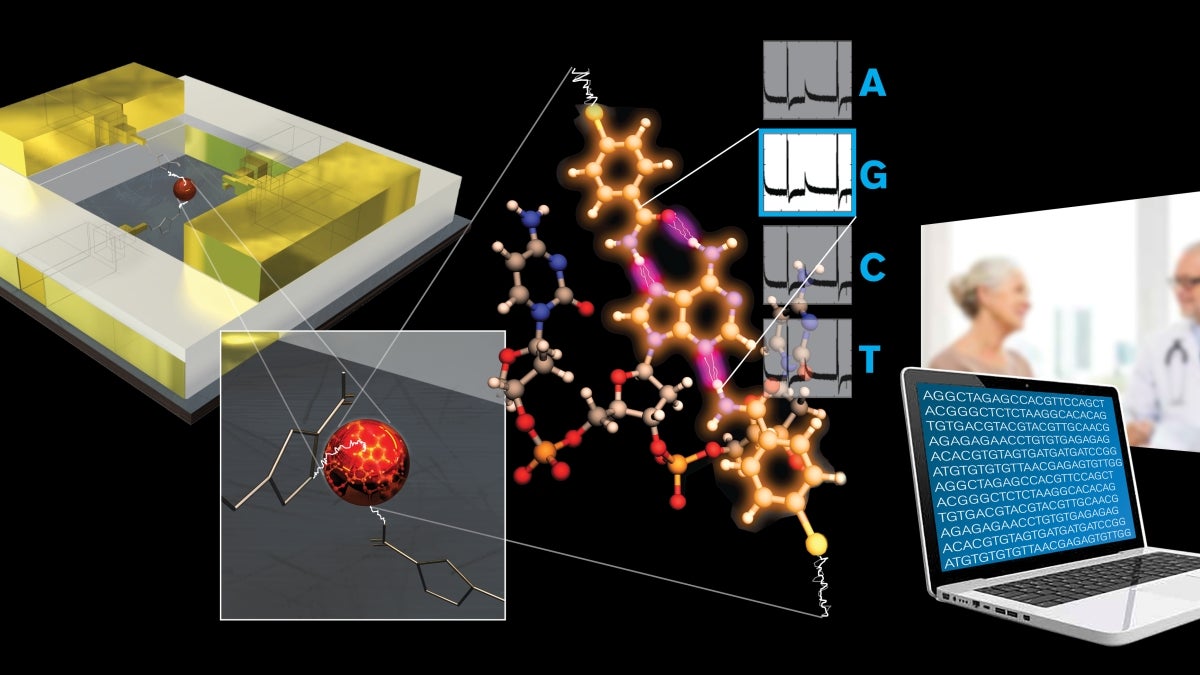
A team of scientists from Arizona State University’s Biodesign Institute and IBM’s T.J. Watson Research Center have developed a prototype DNA reader that could make whole genome profiling an everyday practice in medicine.
"Our goal is to put cheap, simple and powerful DNA and protein diagnostic devices into every single doctor's office," said Stuart Lindsay, an ASU physics professor and director of Biodesign’s Center for Single Molecule Biophysics.
Such technology could help usher in the age of personalized medicine, where information from an individual’s complete DNA and protein profiles could be used to design treatments specific to their individual makeup.
Using tools where biology and physics expertise meet the manufacturing know-how of the semiconductor industry, the team, led by ASU’s Stuart Lindsay and IBM’s Yann Astier, has been developing a device which could make reading an individual’s whole DNA profile, or genome, as easy as passing supermarket goods through a checkout scanner. The first step in doing this is to make a “reading head” that identified single DNA bases as they pass it.
If successful, Lindsay hopes to turn the science of the infinitesimally small (called nanotechnology) into successful products. The ASU group is collaborating with Roche on DNA sequencing while an ASU spinout (Recognition Analytix) hopes to develop a way to sequence single protein molecules.
Such game-changing technology is needed to make genome sequencing a reality. The current hurdle is to do so for less than $1,000, an amount for which insurance companies are more likely to provide reimbursement.
In their latest research breakthrough, the team fashioned a tiny DNA-reading device, thousands of times smaller than the width of a single human hair. The device is sensitive enough to distinguish the individual chemical bases of DNA (known by their abbreviated letters of A, C, T or G) when they are pumped past the reading head.
Proof-of-concept was demonstrated by using solutions of the individual DNA bases, which gave clear signals sensitive enough to detect tiny amounts of DNA (nanomolar concentrations), even better than today’s state-of-the-art, so-called next-generation DNA sequencing technology.
Making the solid-state device is just like making a sandwich, except with ultra high-tech semiconductor tools used to slice and stack the atomic-sized layers of meats and cheeses like the butcher shop’s block. The secret is to slice and stack the layers just so, to turn the chemical information of the DNA into a change in the electrical signal.
First, they made a “sandwich” composed of two metal electrodes separated by a two-nanometer-thick insulating layer (a single nanometer is 10,000 times smaller than a human hair), made by using a semiconductor technology called atomic layer deposition.
Then a hole is cut through the sandwich: DNA bases inside the hole are read as they pass the gap between the metal layers.
“The technology we’ve developed might just be the first big step in building a single-molecule sequencing device based on ordinary computer chip technology,” said Lindsay.
“Previous attempts to make tunnel junctions for reading DNA had one electrode facing another across a small gap between the electrodes, and the gaps had to be adjusted by hand," he added. "This made it impossible to use computer chip manufacturing methods to make devices.
“Our approach of defining the gap using a thin layer of dielectric (insulating) material between the electrodes and exposing this gap by drilling a hole through the layers is much easier. What is more, the recognition tunneling technology we have developed allows us to make a relatively large gap (of two nanometers) compared to the much smaller gaps required previously for tunnel current read-out (which were less than a single nanometer wide). The ability to use larger gaps for tunneling makes the manufacture of the device much easier and gives DNA molecules room to pass the electrodes.”
Specifically, when a current is passed through the nanopore, as the DNA passes through, it causes a spike in the current unique to each chemical base (A, C, T or G) within the DNA molecule. A few more modifications are made to polish and finish the device manufacturing.
The team encountered considerable device-to-device variation, so calibration will be needed to make the technology more robust. And the final big step – of reducing the diameter of the hole through the device to that of a single DNA molecule – has yet to be taken.
But overall, the research team has developed a scalable manufacturing process to make a device that can work reliably for hours at a time, identifying each of the DNA chemical bases while flowing through the two-nanometer gap.
The research team is also working on modifying the technique to read other single molecules, which could be used in an important technology for drug development.
The latest developments could also bring in big business for ASU. Lindsay, dubbed a “serial entrepreneur” by the media, has a new spinout venture, called Recognition Analytix, that hopes to follow the success of Molecular Imaging Corp, a similar instrument company he co-founded in 1993 and sold to Agilent Technologies in 2005.
The research was funded by the National Institutes of Health’s National Human Genome Research Institute, Roche, and published in the journal ACS Nano.