Effective diagnostics, therapies and treatments for diseases and infections could increasingly involve reengineering the body’s internal biomechanisms at their most basic chemical and molecular foundations.
Fulton Schools Associate Professor Xiao Wang. Photo by Jessica Hochreiter/ASU
Growing knowledge about the body’s biological processes is increasing the possibilities for restoring human health, said Xiao Wang, an associate professor of biomedical engineering in Arizona State University’s Ira A. Fulton Schools of Engineering. He and a team of researchers are exploring ways to trigger and control cell differentiation and transition to unlock properties that may change bioengineers’ approach to diagnostics, vaccine development and therapeutic treatments.
Recent research led by Wang and Alexander Green, an assistant professor of biomedical engineering at Boston University, reveals more about the potential for designing small add-on structures for biomolecules that can enhance their properties.
“There could be new and better kinds of applications for diagnostics, therapeutics and treatments, and for genome engineering,” Wang said. “These could be big contributions to biomedicine.”
The details about what the research may yield appear in the paper "Predictable control of RNA lifetime using engineered degradation-tuning RNAs," published this week in the research journal Nature Chemical Biology.
Wang and Green’s focus is on messenger RNA, or mRNA, which carries genetic information from DNA, the molecule that contains the genetic blueprint needed to develop and maintain organisms — including humans.
Alexander Green, assistant professor of biomedical engineering at Boston University and adjunct professor in ASU's School of Molecular Sciences. Photo courtesy of ASU's Biodesign Institute
Within cells, mRNA transmits messages from DNA to the protein-producing ribosomes, informing them of which proteins need to be synthesized at a given time. While DNA’s status as the information repository of the cell means that it is very stable, mRNA’s message-carrying role means it rapidly degrades.
This degradation has made it harder to implement RNA-based therapies and diagnostics.
Wang, Green and their research team are devising methods of controlling degradation to produce predictable, precise and stable results. The new research paper describes how they are attempting to fine-tune the speed of mRNA degradation to boost the ability to perform biotechnological functions. To do this, they have identified specific RNA structural features to build a library of RNA components called degradation-tuning RNAs, or dtRNAs.
Attaching the dtRNAs to an RNA of interest through genetic engineering enables them to increase or decrease the RNA’s degradation rate, and fine-tune gene expression levels in vivo and in vitro — either inside a living organism or in a laboratory setting.
“We found that dtRNAs could be used with a variety of different types of RNAs and modify gene expression levels over a very wide range. These capabilities can increase the speed and sensitivity of medical diagnostics and give us better control over cell function,” said Green, who was an assistant professor in ASU’s Biodesign Institute and School of Molecular Sciences from 2015 to 2020 and is currently an adjunct professor with the school.
One of the more impactful results of these refining processes could be the development of mRNA-based vaccines that would be especially effective against viruses, Wang said.
“We can actually engineer the structure of RNA molecules in faster and more systematic ways that make them more efficient in how they behave,” he said.
These behavior changes will inform how effective Wang and Green’s bioengineering process will be at boosting the efficacy of diagnostics, vaccines, therapies and treatments.
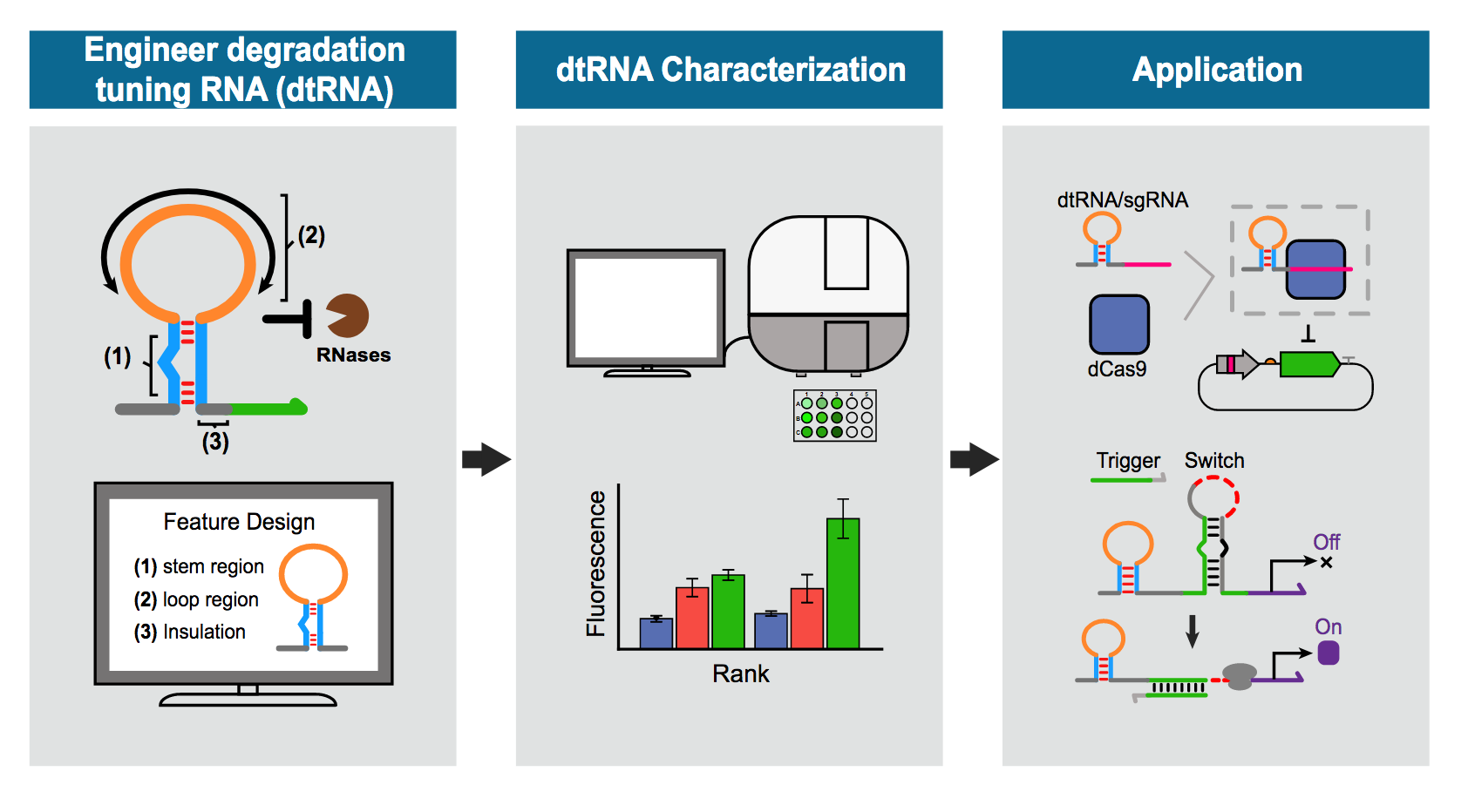
The above graphic provides schematics for a process central to research led by Xiao Wang and Alexander Green to find new ways to restore human health through re-engineering the body’s biomechanisms. In the left and middle panels, the illustration shows the engineering of degradation tuning RNAs, or dtRNAs, by characterizing their structural features. Researchers ranked the entire library of the dtRNAs based on their performance against the RNA degradation process performed by ribonucleases in prokaryotic cells. The right panel shows that researchers tested the potential of these characterized dtRNAs for various biotechnological applications such as regulating gene expression using the gene editing tool CRISPR/Cas9 system or for viral diagnostics by a dtRNA-toehold switch hybrid sensor. Illustration by the Xiao Wang research group
Wang and Green’s research team includes doctoral students Qi Zhang, Duo Ma, Kylie Standage-Beier and Xingwen Chen at ASU, and doctoral student Kaiyue Wu at Boston University. Former ASU doctoral student Fuqing Wu, now at the Massachusetts Institute of Technology, contributed to earlier research that helped lead to the recent discoveries.
Funding for research described in the paper has come from the Arizona Department of Health Services, the Arizona Biomedical Research Centre, the U.S. Department of Health & Human Services, the National Institutes of Health and the NIH Director’s New Innovator Award, the National Science Foundation, the Bill and Melinda Gates Foundation and Arizona State University.
Top image courtesy of Shutterstock
More Science and technology

Real AI solutions helping people now
AI may be everywhere, but its impacts on day-to-day life are more surprising — and beneficial — than you might realize. From scanning thousands of images to prevent crop shortages to creating …

ASU forges strategic partnership to solve the mystery of planet formation
Astronomers have long grappled with the question, “How do planets form?” A new collaboration among Arizona State University, Michigan State University and Lawrence Livermore National Laboratory will…

AI for AZ: ABOR funds new tools for state emergency response
A huge wildfire rages in the wilderness of Arizona’s White Mountains. The blaze scorches asphalt and damages area bridges, causing traffic accidents when residents attempt to evacuate. As hospitals…